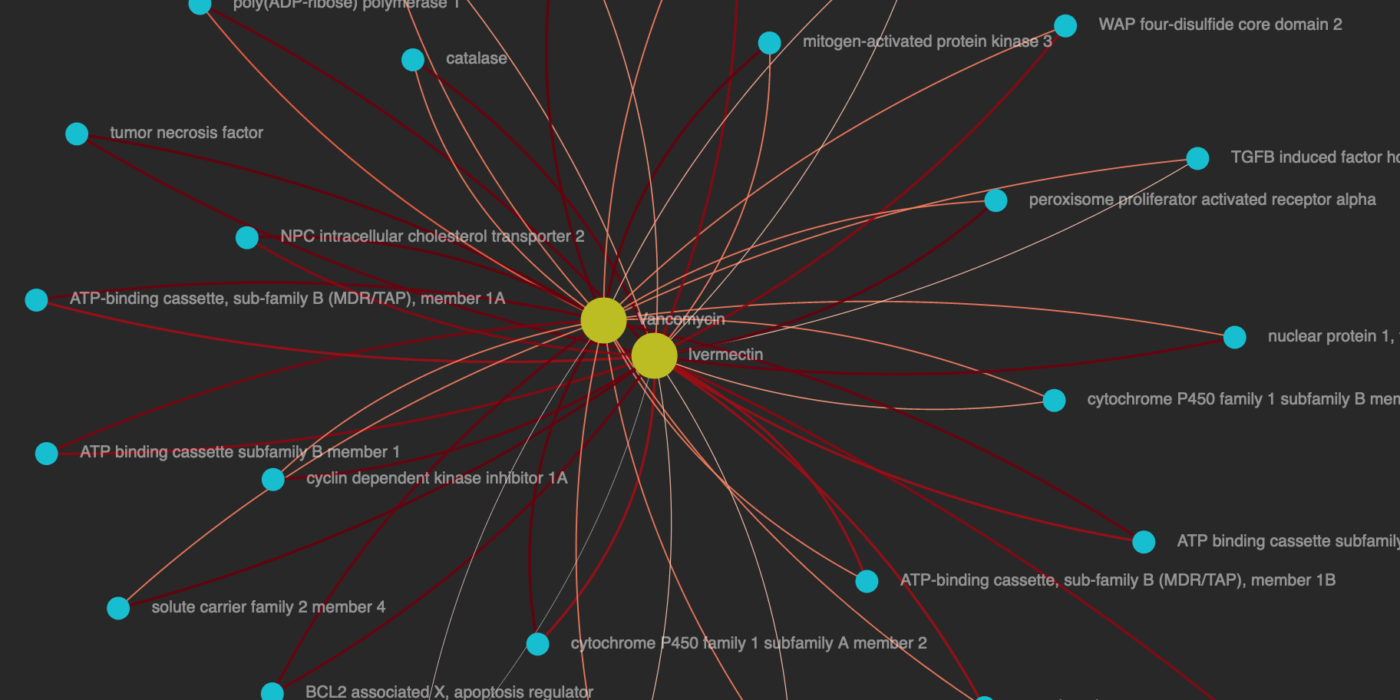
This project builds upon work done by the UIUC Blender Lab. The Blender Lab extracted Knowledge Graphs from analyzing scientific papers about coronavirus. These graphs create connections between chemicals, genes, diseases, etc. that are discussed in these papers.
To take this work a step further, we wanted to make these knowledge graphs useful for scientific researchers during the early days of COVID-19. For example: How could researchers easily see the connections between two pharmaceuticals and make a quick decision about whether to research a particular concept further?
The idea for this project was to utilizing existing in-house graphing technology to enrich these graphs. We used quite a few techniques to enhance the edge (or relationship) weight of two concepts:
- Boltzman Citation Weight – this increases edge weight based on the # of citations, using a Boltzmann sigmoidal equation
- Publication Date Weight – papers that are newer would be weighted higher
- H-Index Weight – it was crucial that researchers were seeing connections that come from reliable authors, especially since the original research dataset included preprint
We also tested extending the functionality of our project by implementing a simple NLP question-answer and summary tool that allowed us to ask simple questions about a particular graph.
Visualizes the strength of the relationship between concepts identified in COVID-19 associated research papers.
In collaboration with: UIUC Blender Lab
- Python
- NetworkX
- AlBERT
- BioPython